Confident genome-wide aneuploidy detection in pre-implantation embryos
Current molecular techniques for pre-implementation genetic screening (PGS) such as traditional fluorescence in situhybridisation (FISH) analysis or more recent bacterial artificial chromosome (BAC)-based CGH arrays have a number of limitations that are successfully overcome using the CytoSure Embryo Screen array. For example, FISH only permits the analysis of a limited number of selected chromosomes, whereas aCGH-based techniques allow the detection of aneuploidies across the whole genome. Furthermore, oligo arrays typically provide lower batch to batch variation, enabling more reliable analysis, plus flexibility in design and content, which further enhances their utility to meet any future research requirements. One of the challenges when working with amplified DNA, particularly when the starting amount is very limited, is that not all of the genome is amplified equally. CytoSure arrays utilise 60-mer oligonucleotide probes, which have been shown to offer higher signal-to-noise ratios and increased specificity and sensitivity than alternative platforms1. The 60-mer oligonucleotide probes used in the design of the CytoSure Embryo Screen array have been selected following sequencing of DNA that has been amplified from single cells using the PicoPLEX® WGA Kit (Rubicon Genomics). This ensures that the array content mirrors the amplified genome. Regions of the genome that are not amplified are not represented, minimising the possibility of obtaining misleading results. For optimum performance, the array can be combined with the CytoSure Genomic DNA Labelling Kit, which offers high signal intensities and an excellent signal-to-noise ratio.
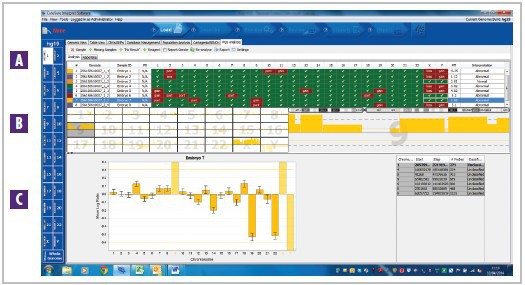
Figure 1: The CytoSure Embryo Screen array in combination with CytoSure Interpret Software enables easy and accurate detection of aberrations. Data obtained from amplified DNA extracted from a single pre-implantation embryo cell. A A summary of multiple sample results, highlighting the gains and losses in each chromosome. Selecting a sample in the table displays it in the B chromosome overview table and C aneuploidy summary plot. The female sample shown here has a gain of chromosome 9*.
"The CytoSure array offers a good alternative to FISH or BAC aCGH for the detection of aneuploidies and chromosomal imbalances in DNA amplified from single embryo cells”. The Center for Human Genetics, University Hospitals Leuven, Belgium.
Optimised data analysis tailored for easy identification of aberrant chromosomes
CytoSure Interpret Software, which accompanies all CytoSure arrays, is a powerful, easy-to-use package for the analysis of aCGH data. A dedicated PGS analysis tab simplifies the detection of aneuploidies and structural aberrations in amplified embryonic samples (Figure 2).

Figure 2: Intuitive data visualisation and interpretation. Amplified DNA isolated from a single embryonic cell was analysed using the CytoSure Embryo Screen array and CytoSure Interpret Software. Aberrations within a single chromosome are calculated using the circular binary segmentation (CBS) algorithm, A the results of which are displayed graphically. Solid blocks indicate aberrations that are considered to be significant. B Tabulated details for each aberration are provided. CClicking on an aberration links to the Genomic View where annotation tracks such as Decipher can aid interpretation.
Multiplex format allows reliable processing of up to fourteen samples on a single slide
Research into the chromosomal content of pre-implantation embryos is typically done with multiple samples. The CytoSure Embryo Screen array can be set-up to suit all laboratory throughput requirements.The low-throughput option requires preparation of eight test and eight reference samples (Figure 3), with the data analysed using standard two-colour hybridisation. Alternatively, for high-throughput requirements,a single-channel approach can be utilised, enabling fourteen samples to be analysed on a single array slide. Cytosure Interpret Software also allows a combination of single-channel and dual-channel analysis on the same slide, enabling fully customisable array set-up. This flexible approach increases processing efficiency and cost-effectiveness as greater multiplexing reduces the number of slides required, minimising both reagents used and handling time.
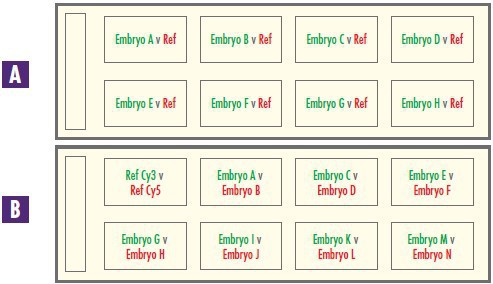
Figure 3: Reliable and cost-effective sample processing. The CytoSure Embryo Screen array can be set-up to run A eight samples or B fourteen samples to suit any throughput requirements. Red text indicates that the sample has been labelled in Cy-5 and green text indicates it has been labelled using Cy-3.
bio-equip.cn







